-Search query
-Search result
Showing all 46 items for (author: radjainia & m)
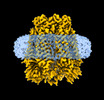
EMDB-15110: 
Cryo-EM structure of mouse Pannexin 1 purified in Salipro nanoparticles
Method: single particle / : Drulyte I

PDB-8a3b: 
Cryo-EM structure of mouse Pannexin 1 purified in Salipro nanoparticles
Method: single particle / : Drulyte I

EMDB-13416: 
Human voltage-gated potassium channel Kv3.1 (apo condition)
Method: single particle / : Chi G, Venkaya S, Singh NK, McKinley G, Mukhopadhyay SMM, Marsden B, MacLean EM, Fernandez-Cid A, Pike ACW, Savva C, Ragan TJ, Burgess-Brown NA, Duerr KL

EMDB-13417: 
Human voltage-gated potassium channel Kv3.1 (with Zn)
Method: single particle / : Chi G, Qian P, Castro-Hartmann P, Venkaya S, Singh NK, McKinley G, Mukhopadhyay SMM, Fernandez-Cid A, Marsden B, MacLean EM, Pike ACW, Sader K, Burgess-Brown NA, Duerr KL

EMDB-13418: 
Human voltage-gated potassium channel Kv3.1 in dimeric state (with Zn)
Method: single particle / : Chi G, Qian P, Castro-Hartmann P, Venkaya S, Singh NK, McKinley G, Mukhopadhyay SMM, Marsden B, MacLean EM, Fernandez-Cid A, Pike ACW, Sader K, Burgess-Brown NA, Duerr KL

EMDB-13419: 
Human voltage-gated potassium channel Kv3.1 (with EDTA)
Method: single particle / : Chi G, Qian P, Castro-Hartmann P, Venkaya S, Singh NK, McKinley G, Mukhopadhyay SMM, Fernandez-Cid A, Pike ACW, Marsden B, MacLean EM, Sader K, Burgess-Brown NA, Duerr KL

PDB-7phh: 
Human voltage-gated potassium channel Kv3.1 (apo condition)
Method: single particle / : Chi G, Venkaya S, Singh NK, McKinley G, Mukhopadhyay SMM, Marsden B, MacLean EM, Fernandez-Cid A, Pike ACW, Savva C, Ragan TJ, Burgess-Brown NA, Duerr KL

PDB-7phi: 
Human voltage-gated potassium channel Kv3.1 (with Zn)
Method: single particle / : Chi G, Qian P, Castro-Hartmann P, Venkaya S, Singh NK, McKinley G, Mukhopadhyay SMM, Fernandez-Cid A, Marsden B, MacLean EM, Pike ACW, Sader K, Burgess-Brown NA, Duerr KL

PDB-7phk: 
Human voltage-gated potassium channel Kv3.1 in dimeric state (with Zn)
Method: single particle / : Chi G, Qian P, Castro-Hartmann P, Venkaya S, Singh NK, McKinley G, Mukhopadhyay SMM, Marsden B, MacLean EM, Fernandez-Cid A, Pike ACW, Sader K, Burgess-Brown NA, Duerr KL

PDB-7phl: 
Human voltage-gated potassium channel Kv3.1 (with EDTA)
Method: single particle / : Chi G, Qian P, Castro-Hartmann P, Venkaya S, Singh NK, McKinley G, Mukhopadhyay SMM, Fernandez-Cid A, Pike ACW, Marsden B, MacLean EM, Sader K, Burgess-Brown NA, Duerr KL

EMDB-20616: 
EM structure of MPEG-1(w.t.) soluble pre-pore
Method: single particle / : Pang SS, Bayly-Jones C

EMDB-20617: 
EM map of MPEG-1 (w.t.) soluble pre-pore complex
Method: single particle / : Pang SS, Bayly-Jones C

EMDB-20619: 
EM map of MPEG-1(L425K, alpha conformation) soluble pre-pore complex
Method: single particle / : Pang SS, Bayly-Jones C

EMDB-20620: 
EM structure of MPEG-1 (L425K, alpha conformation) soluble pre-pore complex
Method: single particle / : Pang SS, Bayly-Jones C

EMDB-20621: 
EM structure of MPEG-1 (L425K, alpha conformation) soluble pre-pore complex
Method: single particle / : Pang SS, Bayly-Jones C

EMDB-20622: 
EM map of MPEG-1(w.t.) pre-pore complex bound to liposome
Method: single particle / : Pang SS, Bayly-Jones C

EMDB-20623: 
EM structure of MPEG-1 (L425K, beta conformation) soluble pre-pore complex
Method: single particle / : Pang SS, Bayly-Jones C

EMDB-20627: 
EM structure of MPEG-1(L425K) pre-pore complex bound to liposome
Method: single particle / : Pang SS, Bayly-Jones C

PDB-6u23: 
EM structure of MPEG-1(w.t.) soluble pre-pore
Method: single particle / : Pang SS, Bayly-Jones C

PDB-6u2j: 
EM structure of MPEG-1 (L425K, alpha conformation) soluble pre-pore complex
Method: single particle / : Pang SS, Bayly-Jones C

PDB-6u2k: 
EM structure of MPEG-1 (L425K, alpha conformation) soluble pre-pore complex
Method: single particle / : Pang SS, Bayly-Jones C

PDB-6u2l: 
EM structure of MPEG-1 (L425K, beta conformation) soluble pre-pore complex
Method: single particle / : Pang SS, Bayly-Jones C

PDB-6u2w: 
EM structure of MPEG-1(L425K) pre-pore complex bound to liposome
Method: single particle / : Pang SS, Bayly-Jones C

EMDB-8978: 
Cryo-EM structure of the active, Gs-protein complexed, human CGRP receptor
Method: single particle / : Liang YL, Khoshouei M, Deganutti G, Glukhova A, Koole C, Peat TS, Radjainia M, Plitzko JM, Baumeister W, Miller LJ, Hay DL, Christopoulos A, Reynolds CA, Wootten D, Sexton PM

PDB-6e3y: 
Cryo-EM structure of the active, Gs-protein complexed, human CGRP receptor
Method: single particle / : Liang YL, Khoshouei M, Deganutti G, Glukhova A, Koole C, Peat TS, Radjainia M, Plitzko JM, Baumeister W, Miller LJ, Hay DL, Christopoulos A, Reynolds CA, Wootten D, Sexton PM

EMDB-7773: 
The X-ray crystal structure of Complement component-9 reveals that the first trans-membrane region acts as a brake on self-assembly
Method: single particle / : Spicer BA, Law RHP, Pang S, Dunstone MA, Whisstock JC

PDB-6dlw: 
Complement component polyC9
Method: single particle / : Dunstone MA, Spicer BA, Law RHP

EMDB-7039: 
3.3 angstrom phase-plate cryo-EM structure of a biased agonist-bound human GLP-1 receptor-Gs complex.
Method: single particle / : Liang YL, Khoshouei M

PDB-6b3j: 
3.3 angstrom phase-plate cryo-EM structure of a biased agonist-bound human GLP-1 receptor-Gs complex
Method: single particle / : Liang YL, Khoshouei M, Glukhova A, Furness SGB, Koole C, Zhao P, Clydesdale L, Thal DM, Radjainia M, Danev R, Baumeister W, Wang MW, Miller LJ, Christopoulos A, Sexton PM, Wootten D

EMDB-3651: 
Cryo-EM asymmetric reconstruction of haemoglobin at 3.6 A determined with the Volta phase plate (subset of particles)
Method: single particle / : Khoshouei M, Radjainia M, Baumeister W, Danev R

EMDB-3650: 
Cryo-EM asymmetric recontsruction of haemoglobin at 3.4 A determined with the Volta phase plate
Method: single particle / : Khoshouei M, Radjainia M, Baumeister W, Danev R

EMDB-8369: 
Methicillin Resistant, Linezolid resistant Staphylococcus aureus 70S ribosome (delta S145 uL3)
Method: single particle / : Belousoff MJ, Lithgow T, Eyal Z, Yonath A, Radjainia M

PDB-5t7v: 
Methicillin Resistant, Linezolid resistant Staphylococcus aureus 70S ribosome (delta S145 uL3)
Method: single particle / : Belousoff MJ, Lithgow T, Eyal Z, Yonath A, Radjainia M

EMDB-8402: 
Methicillin sensitive Staphylococcus aureus 70S ribosome
Method: single particle / : Eyal Z, Ahmed T, Belousoff N, Mishra S, Matzov D, Bashan A, Zimmerman E, Lithgow T, Bhushan S, Yonath A

PDB-5tcu: 
Methicillin sensitive Staphylococcus aureus 70S ribosome
Method: single particle / : Eyal Z, Ahmed T, Belousoff N, Mishra S, Matzov D, Bashan A, Zimmerman E, Lithgow T, Bhushan S, Yonath A

EMDB-8623: 
Volta phase plate cryo-electron microscopy structure of a calcitonin receptor-heterotrimeric Gs protein complex
Method: single particle / : Liang YL, Khoshouei M, Radjainia M, Zhang Y, Glukhova A, Tarrasch J, Thal DM, Furness SGB, Christopoulos G, Coudrat T, Danev R, Baumeister W, Miller LJ, Christopoulos A, Kobilka BK, Wootten D, Skiniotis G, Sexton PM

PDB-5uz7: 
Volta phase plate cryo-electron microscopy structure of a calcitonin receptor-heterotrimeric Gs protein complex
Method: single particle / : Liang YL, Khoshouei M, Radjainia M, Zhang Y, Glukhova A, Tarrasch J, Thal DM, Furness SGB, Christopoulos G, Coudrat T, Danev R, Baumeister W, Miller LJ, Christopoulos A, Kobilka BK, Wootten D, Skiniotis G, Sexton PM

PDB-5ni1: 
CryoEM structure of haemoglobin at 3.2 A determined with the Volta phase plate
Method: single particle / : Khoshouei M, Radjainia M, Bunker R, Baumeister W, Danev R

EMDB-3488: 
CryoEM structure of haemoglobin at 3.2 A determined with the Volta phase plate
Method: single particle / : Khoshouei M, Radjainia R, Baumeister W, Danev R

EMDB-8306: 
CryoEM structure of a beak and feather disease virus-like particle encapsidating ssDNA
Method: single particle / : Hardy JM, Sarker S, Radjainia M, Raidal SR, Forwood JK, Coulibaly F

EMDB-3414: 
Structures of human peroxiredoxin 3 suggest self-chaperoning assembly that maintains catalytic state
Method: helical / : Yewdall AN, Venugopal HP, Desfosses A, Abrishami V, Yosaatmadja Y, Hampton MB, Gerrard JA, Goldstone D, Mitra AK, Mazdak Radjainia M

EMDB-3233: 
Volta phase plate cryo-EM of the small protein complex Prx3
Method: single particle / : Khoshouei M, Radjainia M, Phillips AJ, Gerrard JA, Mitra AK, Plitzko JM, Baumeister W, Danev R

EMDB-6309: 
Cryo-EM structure of human peroxiredoxin-3 filament reveals the assembly of a putative chaperone
Method: single particle / : Radjainia M, Venugopal HP, Desfosses A, Phillips AJ, Yewdall NA, Hampton MB, Gerrard JA, Mitra AK

EMDB-5215: 
Anthrax toxin PA63 in complex with 1G3
Method: single particle / : Radjainia M, Hyun J, Leysath CE, Leppla SH, Mitra AK

PDB-2x8q: 
Cryo-EM 3D model of the icosahedral particle composed of Rous sarcoma virus capsid protein pentamers
Method: single particle / : K Hyun J, Radjainia M, Kingston RL, Mitra AK

EMDB-1710: 
Cryo-EM 3D model of the icosahedral particle composed of Rous sarcoma virus capsid protein pentamers
Method: single particle / : Hyun JK, Radjainia M, Kingston RL, Mitra AK
 Movie
Movie Controller
Controller Structure viewers
Structure viewers About EMN search
About EMN search



 wwPDB to switch to version 3 of the EMDB data model
wwPDB to switch to version 3 of the EMDB data model
